Bark beetles are small insects belonging to the subfamily Scolytinae and are among the most important forest pests. By developing under the bark or within the wood of trees, these beetles damage tree tissues, leading to a deterioration of tree health and, in some cases, death. Although they are essential to the proper functioning of forest ecosystems, outbreak episodes of certain species can have dramatic socio-economic impacts on the forestry sector. In Europe, the most recent outbreaks (2018–2021) of the spruce bark beetle, Ips typographus, resulted in the death of hundreds of millions of cubic metres of spruce.
With international trade, new bark beetle species from other regions of the world are becoming established in our forests. This represents an additional threat, as these species may attack trees that are not adapted to them.
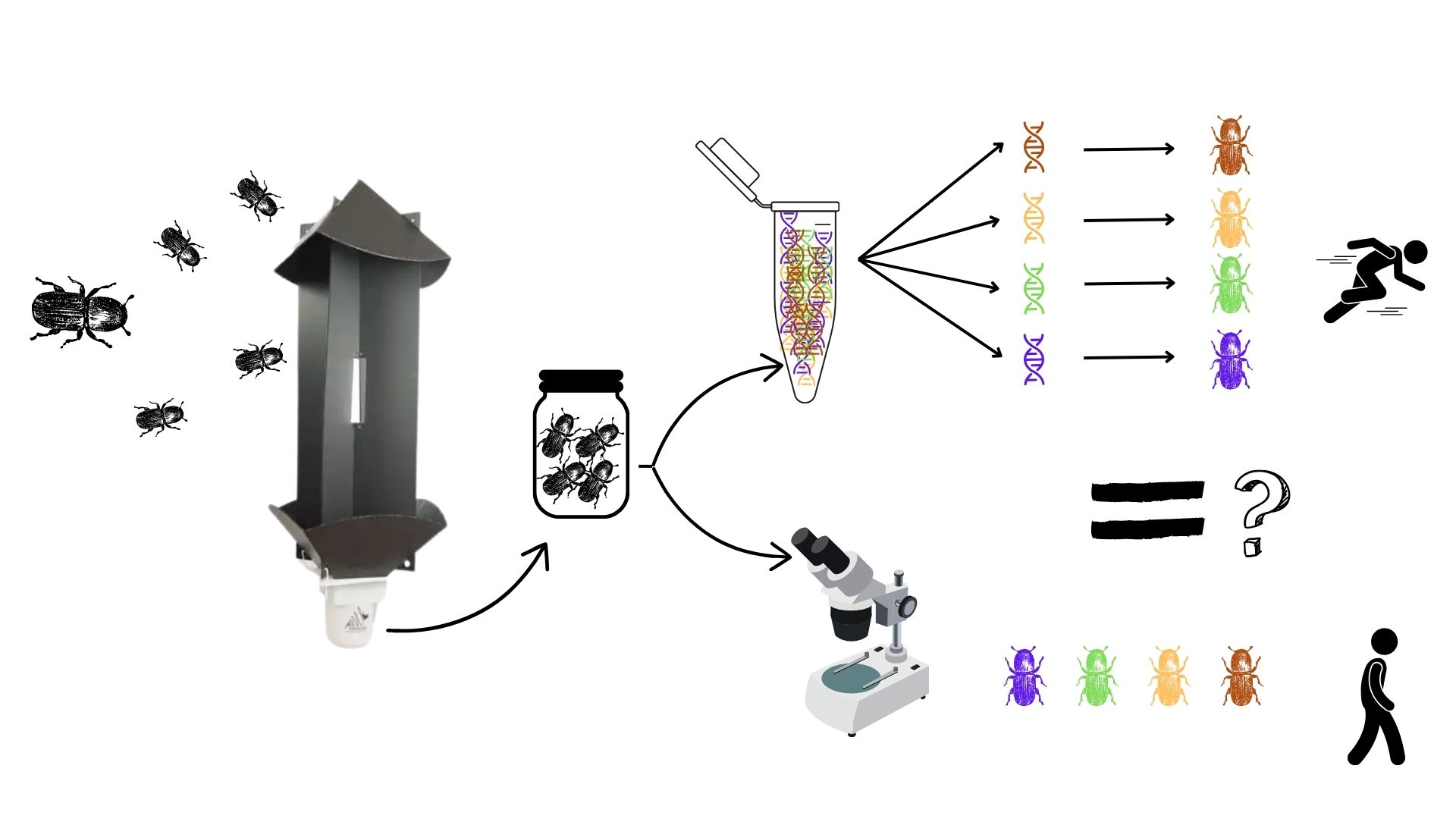
To limit these risks, organisations such as the Federal Agency for the Safety of the Food Chain (FASFC) have implemented surveillance systems. Traditionally, this surveillance relies on the installation of traps combined with morphological identification of the intercepted bark beetles. The identification phase is particularly laborious and requires a high level of expertise to reliably identify all collected specimens. In this context, the use of new molecular tools capable of directly identifying a large number of species within a sample would greatly facilitate the surveillance of non-native bark beetles.
This is where the METASCOL project comes in. Its aim is to test a new identification method known as DNA metabarcoding.
DNA barcoding is a method that allows a species to be identified by reading a short fragment of its DNA and comparing it to a reference database.
DNA metabarcoding works in the same way, but uses modern sequencing techniques to analyse the DNA of multiple organisms simultaneously. Thus, within a single sample (for example, a small amount of soil, water or captured insects), it is possible to detect and identify many different species at the same time.
|
This technique makes it possible to rapidly identify numerous different species from a single sample using their DNA. The objective is to compare the results obtained with this method to those from traditional identification, in order to assess whether it is more efficient and reliable for monitoring bark beetles.
This three-year project (2025–2028), funded by the Federal Public Service for Public Health, will be carried out collaboratively by the entomology laboratories of the Institute for Agricultural, Fisheries and Food Research (ILVO) and the Walloon Agricultural Research Centre (CRA-W). METASCOL logically follows on from a previous project, SCOLIBE, which aimed to identify the non-native bark beetle species posing the greatest risk to Belgium and to optimise trapping devices (location, trap types and attractants) for their surveillance.
If DNA metabarcoding proves capable of reliably detecting the bark beetles present in a sample, it would greatly facilitate the monitoring of non-native species. More broadly, this project contributes to the preservation of Belgian forest resources from damage that may be caused by exotic bark beetles.